Recently, the Institute of Vegetables and Flowers constructed the Solanaceae inter-species pan-genome based on 46 high-quality genomes of Solanaceae species. The study explored the genomic variation characteristics and mechanisms at an inter-species evolutionary scale, as well as the functional differentiation of polyploidy genes. These findings provide important insights for comparative evolutionary studies and functional gene mining and breeding applications in Solanaceae species. The results were published in Plant Communications under the title “SolPGD: Solanaceae Pan-genomes Reveal Extensive Fractionation and Functional Innovation of Duplicated Genes.”
Solanaceae plants, including model crops like tomato, pepper, potato, eggplant, and tobacco, as well as orphan crops like wolfberry, groundcherry, and pepino, play a significant role in agriculture. While research on model crops has made substantial progress, studies on orphan crops lag behind, with their breeding potential largely untapped. This study established the Solanaceae inter-species pan-genome to analyze gene retention and loss patterns, providing essential information for leveraging model crop research to advance other species.
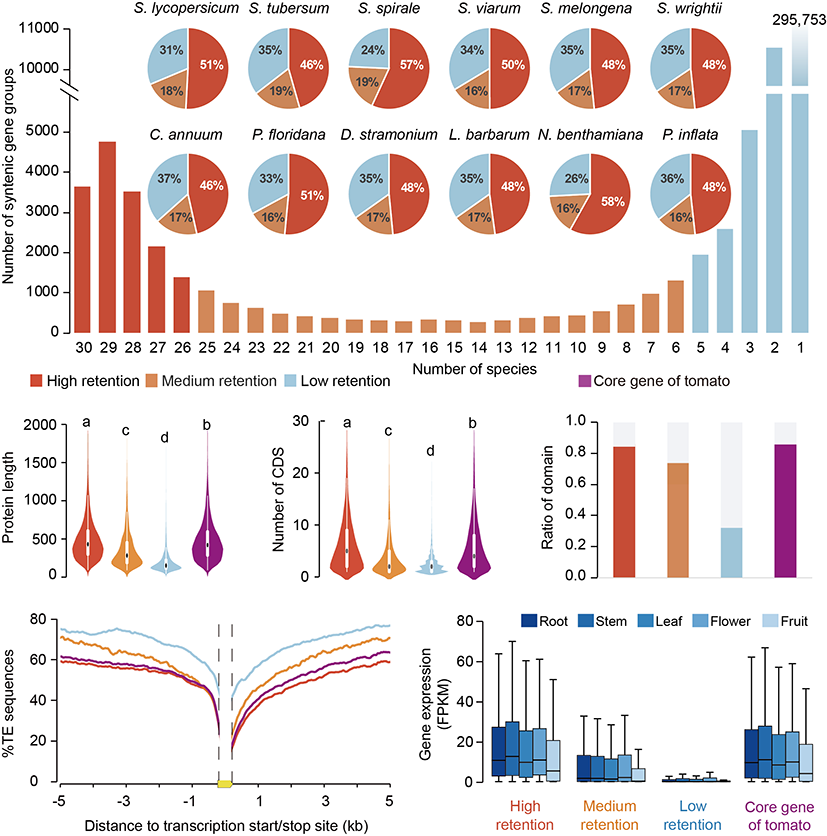
The research was based on 46 high-quality genomes (assembled using third-generation sequencing data) selected from 208 Solanaceae plant genomes, with a strict filtering process based on species divergence time (no less than 6 million years) and genome completeness. This set of genomes, covering key species within the family, lays a foundation for cross-species comparative analysis. Through synteny analysis, 43,395 syntenic gene families were identified. The study proposed a classification system for gene families suitable for inter-species pan-genome analysis: high retention genes (shared by >80% of species), medium retention genes (shared by 20%-80% of species), and low retention genes (shared by <20% of species). High retention genes were found to have longer protein sequences, higher functional conservation, and expression levels, indicating their vital role in the basic biological functions of Solanaceae plants.
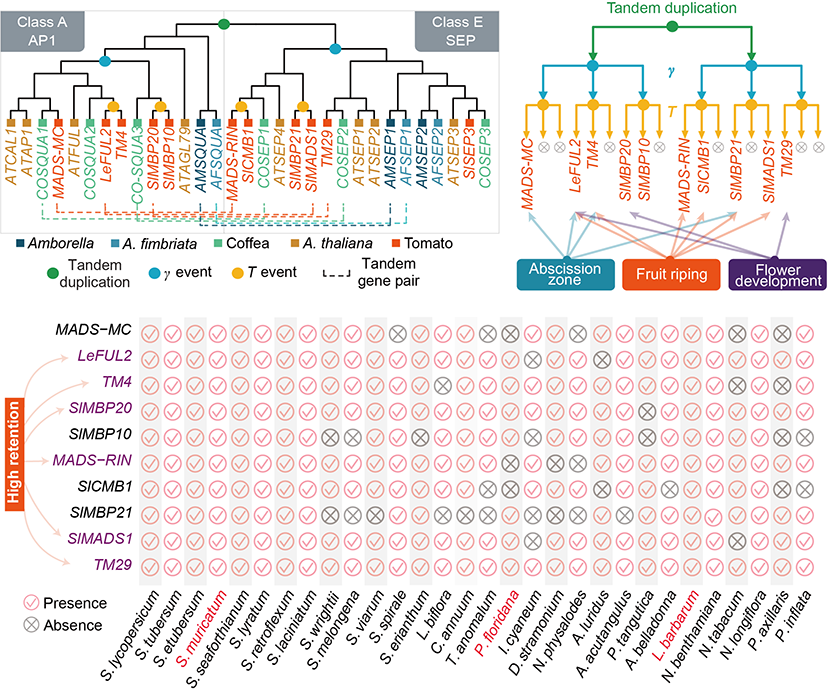
Further analysis with 9 diploid genomes from non-Solanaceae species provided a comprehensive understanding of the Solanaceae-specific whole-genome ancient triplication event (T event) that occurred 65 million years ago and identified multi-copy genes generated from this event. This gene set is valuable for tracing the origins and differentiation of key functional genes in Solanaceae plants. For example, in the flower development ABCDE gene model, it was found that all genes in the Solanaceae A and E gene families originated from an ancient tandem duplication event, followed by two whole-genome ancient triplication events (the γ event, shared by all eudicots, and the T event specific to Solanaceae) and subsequent gene losses, leading to 10 homologous genes. These genes are highly conserved across Solanaceae species. Moreover, several high-retention genes exhibited similar gene expression patterns in model crops (tomato, pepper) and orphan crops (wolfberry, groundcherry), suggesting functional conservation across different species.
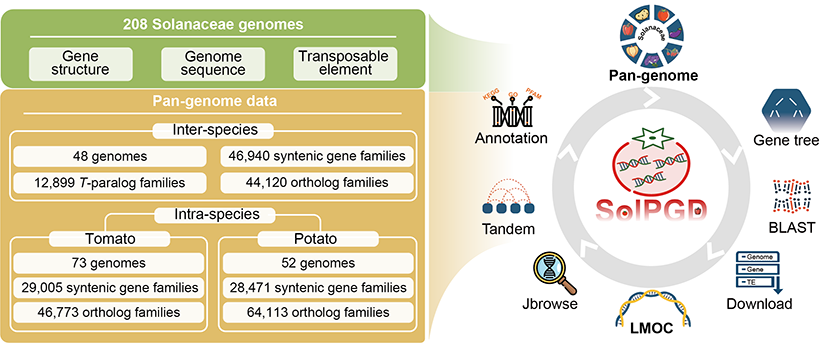
To better serve researchers in this field, the team developed the Solanaceae Pan-genome Database (SolPGD, http://www.bioinformaticslab.cn/SolPGD), which offers a user-friendly platform for querying Solanaceae genome data, gene family homology, and gene function similarity comparisons between orphan and model crops. This resource supports scientific research and breeding applications in Solanaceae species.
In summary, this study provides a new perspective on genome evolution and gene functional differentiation in Solanaceae plants through inter-species pan-genome construction at the family level, offering an important database platform for comparative genomics and gene mining for researchers in the field.
Original Link: https://www.sciencedirect.com/science/article/pii/S2590346224006527?via%3Dihub