Spinach originated from Persia (modern-day Iran), also known as Persian grass or red-rooted vegetable, and was introduced to China as a tribute from Nepal during the Tang Dynasty. Spinach (Spinacia oleracea L.) has two different types of wild species, S. turkestanica and S. tetrandra. Wild materials often possess strong adaptability and stress resistance, making them important resources for crop genetic improvement. However, the genomic information of the two wild types of spinach is unclear, hindering further utilization of spinach wild species. Therefore, deciphering the genome sequences of spinach wild species and identifying important genes related to domestication traits lay the foundation for breeding high-quality spinach varieties.
Recently, the Spinach Genetic Breeding Research Team led by Qian Wei from the Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences, published a research paper titled 'Insights into spinach domestication from genomes sequences of two wild spinach progenitors, Spinacia turkestanica and Spinacia tetrandra' in the internationally renowned journal New Phytologist. The study assembled the genomes of two wild spinach progenitors and elucidated their important domestication traits.

The research integrated second-generation, third-generation, and Hi-C sequencing to successfully assemble two high-quality reference genomes of wild spinach (Figure 1). Based on single-copy genes, it was confirmed that Spinacia turkestanica is the direct ancestor of cultivated spinach, with an estimated divergence time of approximately 0.8 million years ago, while Spinacia tetrandra diverged even earlier, approximately 6.3 million years ago.
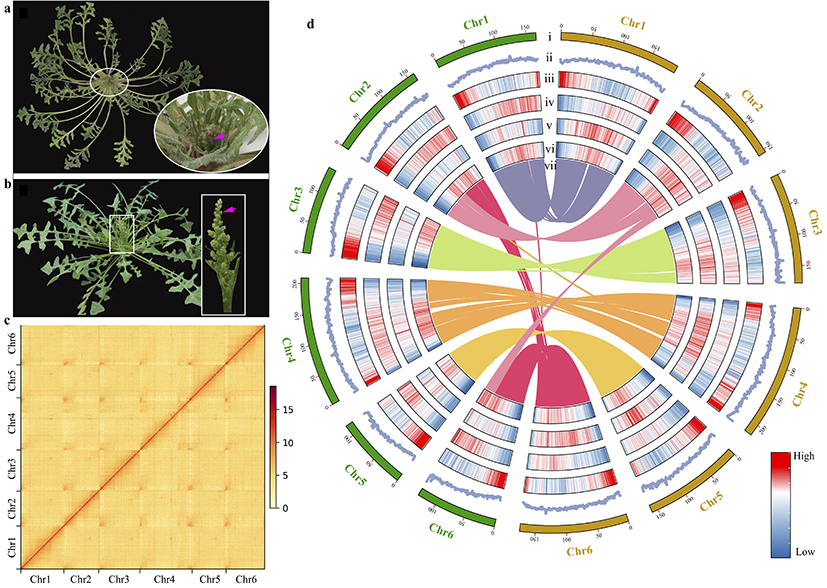
Figure 1 Morphology of individuals and inflorescences of the two wild spinach species, illustrating the differences between them, and the species’ genome features.
The team led by Qian Wei previously assembled a high-quality reference genome of cultivated spinach (She et al., 2023, Plant Physiology) and combined it with genetic linkage maps to discover that the telomeric regions of each autosomal end and the central region of the sex chromosome (chromosome 4) are enriched with abundant repetitive sequences and exhibit lower recombination rates. This phenomenon is also observed in the genomes of the two wild species, where these regions may serve as pericentromeric regions, leading to the accumulation of repetitive sequences. Within these regions, nucleotide polymorphism decreases, linkage disequilibrium (LD) values increase, the rate of LD decay decreases, and recombination rates decrease (Figure 2).
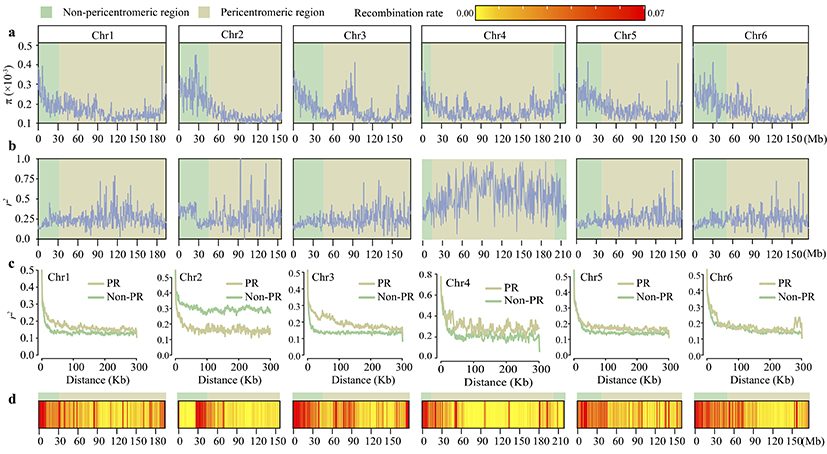
Figure 2 Distribution of nucleotide diversity (p), recombination rate, and linkage disequilibrium (LD) values.
Based on the high-quality reference genome of spinach and 94 accessions of spinach germplasm resources, we identified 982,999 high-quality SNPs and 71,115 structural variations. Using structural variations and population data, we identified six introgression regions in the genome. Among them, the spinach downy mildew resistance loci RPF1–RPF3 and RPF12 are all located within the introgression region of Chr1 0.5–1.5 Mb, indicating that the resistance of cultivated spinach to downy mildew is derived from introgression from wild species. Additionally, we identified numerous domestication-related genes, mainly related to photoperiod and disease resistance. For example, a 620-bp sequence insertion was found downstream of SpCUC3, which is under selection pressure in cultivated spinach, leading to reduced expression of this gene in cultivated varieties. Previous studies have shown that silencing the CUC3 gene can smooth the leaf margin from serrated to smooth. While the leaf margin of spinach wild species appears serrated, cultivated varieties mostly exhibit smooth margins, suggesting that SpCUC3 may regulate the leaf margin of spinach.
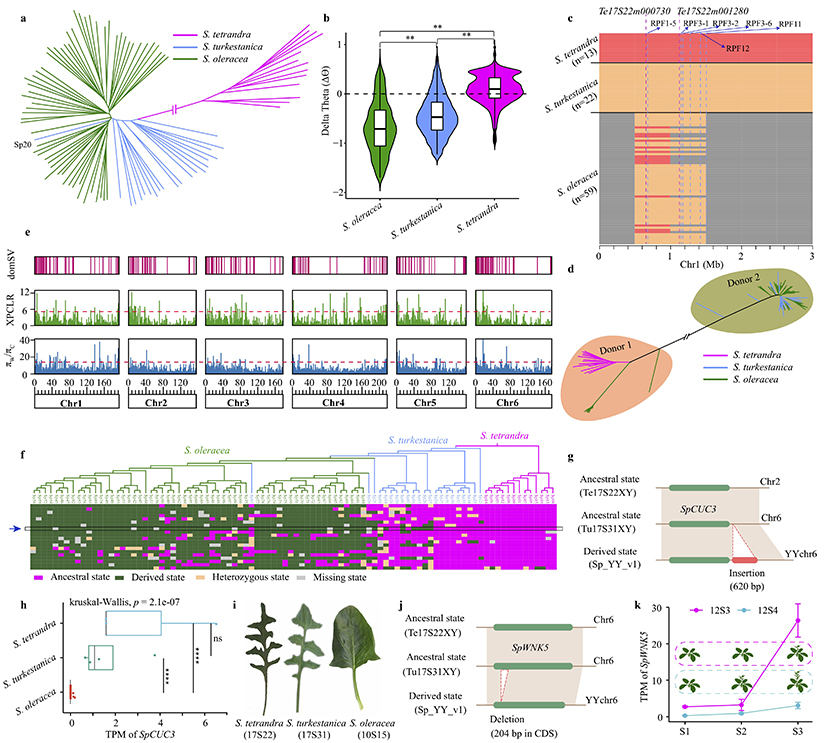
Figure 3 Phylogeny, introgression, and domestication in cultivated spinach and its wild relative specie
In summary, this study elucidated the genomic information of two wild spinach species, identified important domestication-related genes, and clarified that the downy mildew resistance loci in cultivated spinach originated from introgression from wild species. These findings provide theoretical guidance for further utilization of wild spinach resources and breeding high-quality spinach varieties.
Assistant Researcher She Hongbing and Researcher Liu Zhiyuan from the Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences, are joint first authors of this paper; Researcher Qian Wei and Researcher Cheng Feng from the Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences, as well as Professor Deborah Charlesworth from the University of Edinburgh, United Kingdom, are joint corresponding authors. This research was supported by the Chinese Academy of Agricultural Sciences Innovation Project (CAAS-ASTIP-IVFCAAS, CAAS-ZDRW202103) and the China Agricultural Research System (CARS-23-A-17).
More information can be found through the link: https://nph.onlinelibrary.wiley.com/doi/epdf/10.1111/nph.19799.