Recently, Prof. Mao Zhenchuan team from the Plant Disease Group of the Institute of Vegetables and Flowers collaborated with Prof. Pierre Abad team from the Institut Sophia Agrobiotech publish an article “A nematode effector Mi2G02 hijacks a host plant trihelix transcription factor for nematode parasitism" in Plant Communications. In this paper, they reported the molecular mechanism by which the Mi2G02 effector protein secreted by Meloidogyne incognita interacted with the host trihelix transcription factor GT-3a to regulate the expression of downstream target genes and promote nematode parasitism.
Plant-parasitic nematodes cause great agricultural losses every year. Root-knot nematodes (RKNs) and cyst nematodes are harmful and economically important crop pathogens. RKNs can infect almost all cultivated plants, especially some vegetable crops such as tomato, cucumber and pepper. With the global climate change, the domestic planting system reform and the rapid development of agricultural mechanization, large-scale and high-value agriculture, the RKNs disease of vegetables has a serious tendency. During the parasitic stages, a large number of effector proteins are secreted into host plant cells by esophagus glands and epidermis, which can degrade plant cell wall, regulate plant immune response, interfere with hormone synthesis and signal transduction, regulate gene expression and cell development. By analyzing the functions of the key effector proteins and host target proteins during RKNs parasitism will provided theoretical basis for finding new targets for controlling RKNs and developing RNA biopesticides.
In this study, host-induced gene silencing and transgenic method were used to demonstrate that the Mi2G02 effector protein of the Meloidogyne incognita was involved in the regulation of root length and the development of nematode feeding cells. Yeast two-hybrid, bimolecular fluorescence complementation, luciferase complementation assay, and co-immunoprecipitation experiments confirmed that Mi2G02 interacted with plant trihelix transcription factor GT-3a in the nucleus, and the nuclear localization signal of Mi2G02, ShKT domain and DNA-binding domain of GT-3a were the key regions for interaction. GT-3a played an important role in plant root development, especially lateral root development, and was also a key plant target for successful RKNs parasitism. GT-3a had transcriptional inhibition function and could specifically bind the promoter sequence of TOZ and RAD23 and suppressed their expression. Further studies showed that Mi2G02 stabilized GT-3a by inhibiting 26S proteasome-mediated protein degradation, and increased the inhibitory effect on TOZ and RAD23 gene expression. In summary, this study reported the molecular mechanism by which Mi2G02, a key effector of nematode parasitism, interacted with the host transcription factor GT-3a, inhibited its degradation and regulates the expression of downstream target genes, thereby promoting nematode to establish a parasitic relationship with its host.
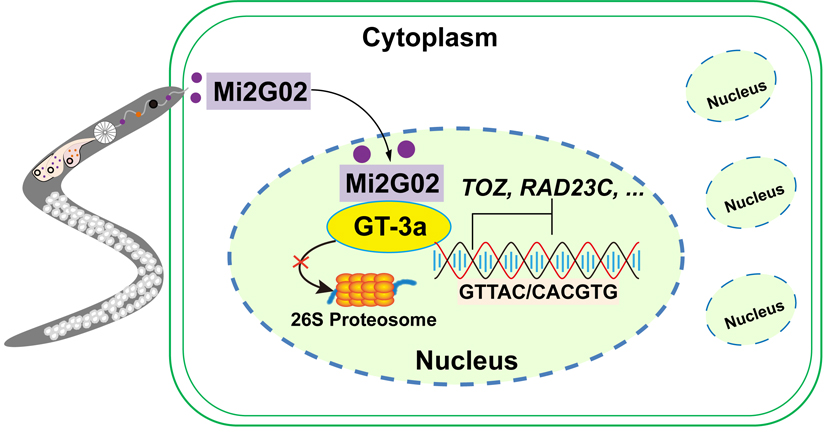
Fig. Model of interaction between Mi2G02 effector protein and host GT-3a to promote nematode parasitism
Zhao Jianlong, a researcher at the Institute of Vegetables and Flowers of the Chinese Academy of Agricultural Sciences, Huang Kaiwei, a master's student, and Liu Rui, a doctoral student, are the co-first authors of this paper. Zhao Jianlong, Mao Zhenchuan, and Michaël Quentin are the co-corresponding authors. Researcher Yang Yuhong, Researcher Ling Jian, Researcher Li Yan, and Researcher Xie Bingyan from the Institute of Vegetables and Flowers of the Chinese Academy of Agricultural Sciences. Professor Jian Heng from China Agricultural University and Professor Pierre Abad and Professor Bruno Favery from the Institut Sophia Agrobiotech participated in this work. This study has been supported by multiple funds, including the Youth Innovation Program of the Chinese Academy of Agricultural Sciences, the National Natural Science Foundation of China, the Beijing Natural Science Foundation, the Innovation Project of the Chinese Academy of Agricultural Sciences, and other fundings.
The article link: https://doi.org/10.1016/j.xplc.2023.100723