Recently, the Vegetable Molecular Design Breeding Innovation Team at the Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences, has developed a proximity-dependent protein labeling system targeting specific DNA sequences (CSPL). This system is applied to efficiently capture proteins bound to specific locations in the plant genome, such as transcription factors interacting with promoters, providing a crucial tool for deciphering the upstream regulatory factors of target genes. The related findings have been published in Nature Plants.

In their research on brassica vegetables, the team discovered that approximately 70% of gene expression changes are associated with structural variations (SVs), with some SV sequences enriched for transcription factor binding sites. While key domestication genes for leafy head formation like ACS and KAN1 were previously located based on SVs (Nature Genetics, 2024), and regulatory genes for head development such as PIF4 were identified (Plant Journal, 2022), the upstream signals, regulatory factors, and underlying molecular mechanisms remained unclear. To delve deeper into the functional mechanisms of regulatory genes controlling traits like head formation, the research team developed a technique for efficiently identifying upstream regulatory factors of target genes.
The interactions between regulatory proteins like transcription factors and their target genomic sites are dynamic, characterized by weak binding affinity, low abundance, and transient duration. Existing experimental techniques, such as chromatin immunoprecipitation, suffer from low identification efficiency or dependence on specific antibodies, limiting related research. Capturing these fleeting key regulatory factors without interfering with the normal life activities of living cells poses a significant challenge in plant gene transcription regulation studies. To address this, the team created the CSPL system by combining a "navigation system" (dCas9) for precise targeting of specific DNA sequences with a "labeler" (TurboID enzyme) for rapid protein tagging. By expressing a dCas9-TurboID fusion protein in plant cells, it precisely targets the desired DNA sequence and biotinylates nearby proteins. Subsequent streptavidin enrichment and mass spectrometry identification then provide information on proteins bound to the target sequence. Using the CSPL system, the team targeted the light-temperature pathway gene PIF4 (involved in regulating head development) and "scanned" its promoter sequences in cabbage, Arabidopsis, and rice. They successfully captured not only reported upstream transcription factors of PIF4 like TCPs and FHY3, as well as epigenetic regulators like ELFs and PWR, but also identified novel transcription factors such as GTL2 and LSH3. The successful application of CSPL in multiple plant species demonstrates its high sensitivity and broad applicability.
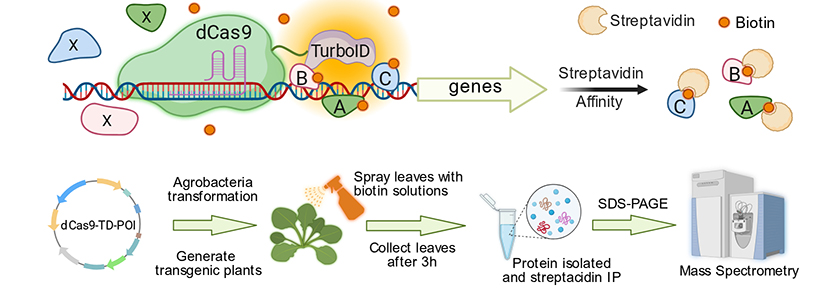
The CSPL technology provides a universal solution for efficiently identifying upstream regulatory factors of target genes across various plant species, different tissues, and under diverse growth conditions. Beyond PIF4, the team has also utilized CSPL to investigate the upstream regulators of cabbage head domestication genes like ACS, aiming to unravel the molecular regulatory mechanisms of leafy head formation.
Associate Researcher Zhang Lei, Associate Researcher Cai Chengcheng, and Master's student Chen Qiujie from the Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences, are the co-first authors of the paper. Researcher Cheng Feng is the corresponding author. This research was supported by the National Major Agricultural Science and Technology Project of China, the National Natural Science Foundation of China, and the Agricultural Science and Technology Innovation Program of the Chinese Academy of Agricultural Sciences.
Article link: https://www.nature.com/articles/s41477-025-02212-5