Recently, the Molecular Design Breeding Team and the Pepper Breeding Team at the Institute of Vegetables and Flowers of the Chinese Academy of Agricultural Sciences collaboratively constructed a high-quality complete genome map of the pepper variety “Zunla-1”. The related findings were published in Plant Communications under the title “The Gap-free Genome of Pepper Reveals the Transposable Element-driven Expansion and Rapid Evolution of Pericentromeres”.

Pepper (Capsicum annuum L.) is a widely cultivated vegetable crop and spice around the world. The pepper genome exceeds 3 Gb in size, making it 3 to 4 times larger than the genomes of other common Solanaceae vegetables such as tomato and potato, and it is highly complex. The cultivated pepper variety “Zunla-1” had its whole genome draft (V2.0) completed in 2014, and it is one of the two most commonly used reference genomes in pepper research. However, due to the limitations of sequencing technology at that time, the continuity and completeness of the Zunla-1_V2.0 genome were relatively low.
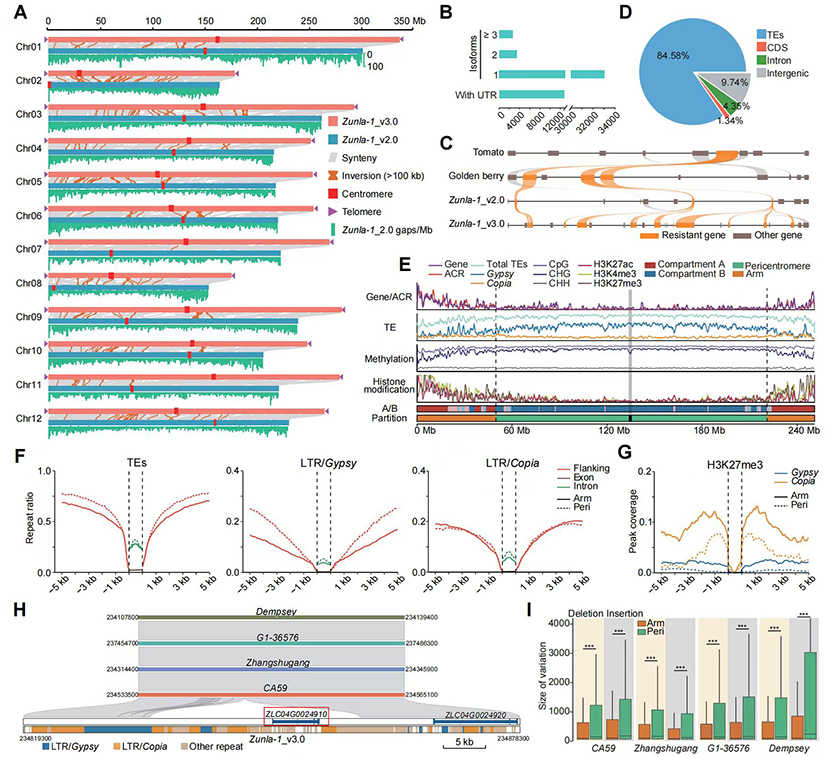
This study utilized HiFi, Ultra-long, and Hi-C sequencing technologies to assemble the “Zunla-1” genome, resulting in a size of 3.08 Gb with a contig N50 of 167.63 Mb, successfully obtaining a complete, the telomere to telomere gap-free genome of pepper (Zunla-1_v3.0). The genome consists of approximately 84% repetitive sequences, with a total of 39,068 genes annotated and a large number of disease resistance gene clusters identified. The centromeres of pepper do not possess typical satellite sequences, and therefore the 12 centromeres were successfully identified based on centromere-specific three-dimensional interaction patterns. By integrating sequence, epigenetic, and three-dimensional genomic characteristics, the chromosomes were categorized into telomeric regions, arm regions, pericentromeric regions, and centromeric regions. The study found that the pericentromeric regions of pepper significantly expanded, exhibiting sparse gene distribution and a high enrichment of Gypsy retrotransposons, while Copia retrotransposons were relatively enriched in the arm regions. This differential distribution may be related to their epigenetic modification patterns. Furthermore, intra-species and inter-species differences were more pronounced in the pericentromeric regions than in other genomic areas, suggesting that repetitive sequences have led to the expansion of the pericentromeres in pepper, promoting high variability in these regions. The reference genome Zunla-1_v3.0 released in this study shows a significant improvement in quality compared to previous versions and will provide efficient support for future functional genomic studies and molecular breeding in pepper (http://www.bioinformaticslab.cn/PepperBase/).
Professor Kang Zhang, PhD student Xiang Wang and Associate Professor Shumin Chen from the Institute of Vegetables and Flowers of the Chinese Academy of Agricultural Sciences are co-first authors of the paper, while Professors Feng Cheng and Lihao Wang are co-corresponding authors. Professor Jue Ruan from the Agricultural Genomics Institute of the Chinese Academy of Agricultural Sciences also participated in the project. This research was supported by the State Key Laboratory of Vegetable Biobreeding, the National Natural Science Foundation, and the Technology Innovation Project and Basal Research Fund of the Chinese Academy of Agricultural Sciences.
Paper link: https://www.sciencedirect.com/science/article/pii/S2590346224005935