Recently, the Vegetable Molecular Design Breeding Innovation Team and the Solanaceae Vegetable Breeding Innovation Team at the Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences (IVF-CAAS), constructed high-quality genomes for wild and cultivated peppers. They analyzed the chromosomal karyotypes, 3D genome structures, and the evolution of gene regulatory elements, uncovering the significant roles of transposable element-driven structural variations and frequent introgressions in shaping key traits of peppers. These findings were published in Nature Plants under the title “Transposon proliferation drives genome architecture and regulatory evolution in wild and domesticated peppers.”

Peppers (Capsicum spp.) are one of the most extensively grown and economically valuable vegetable crops in China. The genus Capsicum consists of over 30 species, including five domesticated species and many wild species, exhibiting rich genetic diversity. However, the evolutionary trajectory of Capsicum remains unclear. Pepper genomes, typically exceeding 3 Gb, are about four times the size of tomato genomes, with high repeat content. Do these repeats influence genome evolution and trait development? Addressing these questions will enhance our understanding of pepper germplasm evolution, trait mechanisms, and genetic innovation.
In this study, researchers assembled 11 high-quality Capsicum genomes using HiFi and ONT sequencing technologies, achieving an average contig N50 of over 230 Mb. The genomes include all five domesticated species (C. annuum, C. baccatum, C. chinense, C. frutescens, and C. pubescens) and five wild species, including the distant relative C. rhomboideum in the Andes region of South America. Among these, the C. rhomboideum genome is the smallest (1.80 Gb), approximately half the size of other pepper genomes. Most genomes have fewer than six gaps, while the genomes of C. rhomboideum and the wild C. annuum accession PM1533 achieved telomere-to-telomere (T2T, gap-free) assemblies, providing a robust foundation for subsequent analyses.
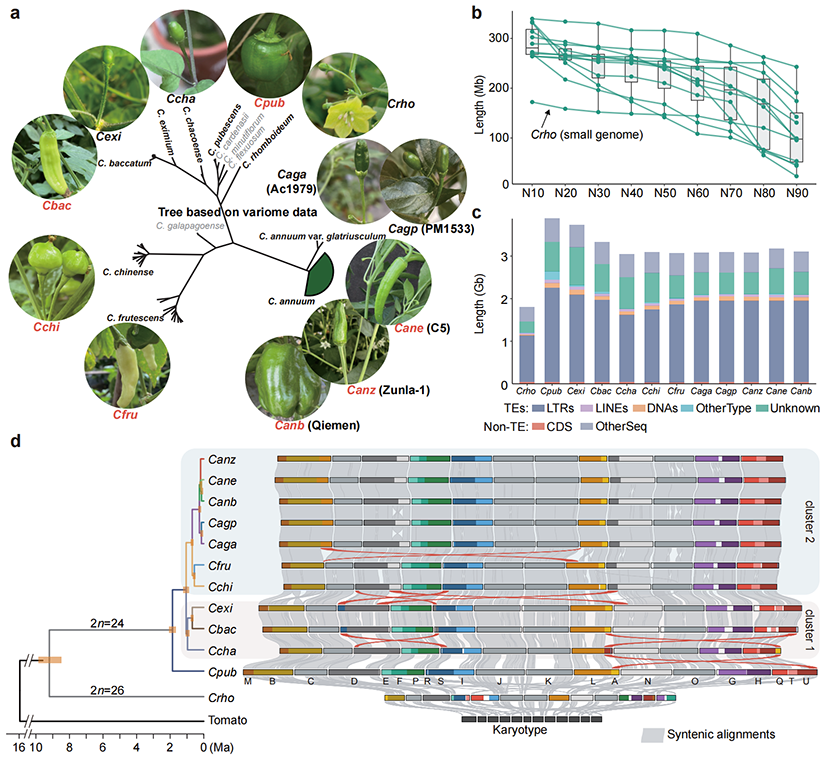
Fig 1. Sequencing and assembly of wild and domesticated pepper genomes
Phylogenetic analyses revealed that C. rhomboideum, representing the Andean wild pepper lineage, was the earliest to diverge, followed by C. pubescens. The remaining pepper species were divided into two major clusters. Based on evolutionary relationships and genome synteny, the ancestral karyotype of Capsicum was reconstructed, confirming that the ancestral Capsicum genome had 12 pairs of chromosomes. The 13 pairs observed in C. rhomboideum resulted from chromosomal rearrangements of the ancestral karyotype, resolving long-standing debates over the ancestral chromosome number in peppers. Comparisons of 3D chromatin compartments among species revealed that chromosomal rearrangements induced significant transitions in the compartments, shedding light on the relationship between chromosomal rearrangements and three-dimensional genome architecture.
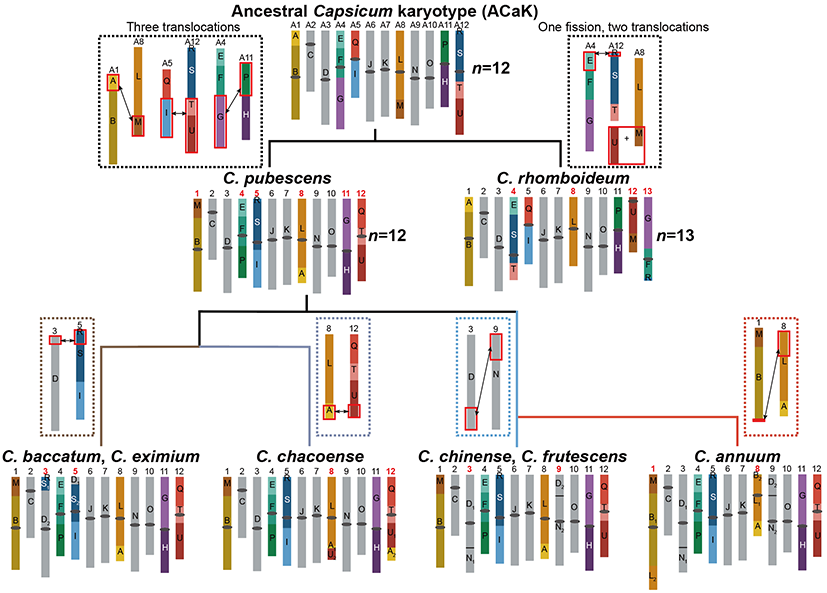
Fig 2. Ancestral karyotype of Capsicum and its evolution during pepper speciation
The study uncovered striking differences in transposable element (TE) accumulation across pepper genomes. Recent bursts of Gypsy-type retrotransposons in species such as C. pubescens and C. baccatum contributed to their relatively larger genome sizes. These TE insertions also generated widespread structural variations (SVs) among genomes. Notably, an 8-kb deletion in the promoter region of the Up gene, which regulates fruit orientation, suppressed its expression in the C. chinense pepper with assembled genome, causing fruit orientation to shift from downward to upward. Similarly, in C. pubescens, a TE insertion in the coding region of the CCS gene, critical for capsanthin synthesis, led to premature termination and a shift in fruit color from red to yellow. These results provide important insights into the genetic mechanisms underlying trait diversity in peppers.
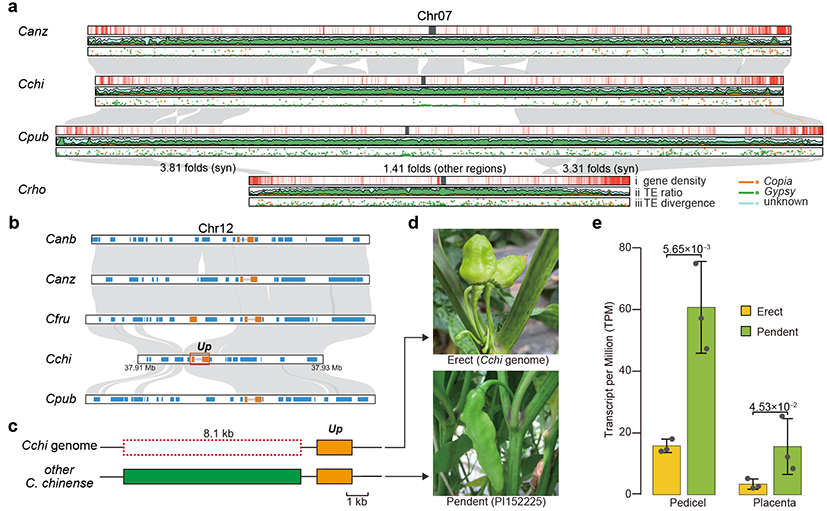
Fig 3. Comparison of Capsicum genomes and variation in Up gene that regulates the fruit orientation
This study constructed the first chromatin accessibility map of the Capsicum genus, revealing the dynamic evolution of regulatory elements. Chromatin accessibility, a key determinant of gene regulation, plays a pivotal role in the “on-and-off” switching of genes. However, the impact of TEs on accessible chromatin regions (ACRs) during pepper genome expansion remains unclear. Comparative analyses across pepper species revealed that both the emergence and decay of ACRs are closely associated with TEs. Species-specific ACRs were significantly enriched in TEs, while ancestral ACRs exhibited a negative correlation between their conservation and TE density. SVs within ACRs further reduced their conservation. Notably, new ACRs near genes such as IQD, a regulator of fruit shape, were associated with its high expression in sweet peppers, suggesting that these new ACRs may have facilitated changes in sweet pepper fruit shape. These findings provide new insights into the dynamic regulation of gene expression in peppers.
Additionally, the study performed deep resequencing of 124 core Capsicum accessions, constructing a high-resolution haploblock map of the pepper population. Analyses revealed extensive interspecific introgressions from C. frutescens into C. annuum, as well as significant haploblock differentiation within the C. annuum population. Late-evolving blocky sweet peppers exhibited interspecific introgressions and intraspecific haploblock divergence. For example, the haploblocks containing the PMMoV resistance gene L showed notable diversity within the C. annuum population, with some sweet pepper varieties acquiring the L3 genotype introgressed from C. chinense peppers to gain PMMoV resistance. Furthermore, the absence of pungency in sweet peppers was linked to stepwise selection and differentiation of haploblocks surrounding the Pun1 locus, a key regulator of capsaicin biosynthesis. These introgressions and haploblock variations greatly enriched the genetic and phenotypic diversity of C. annuum.
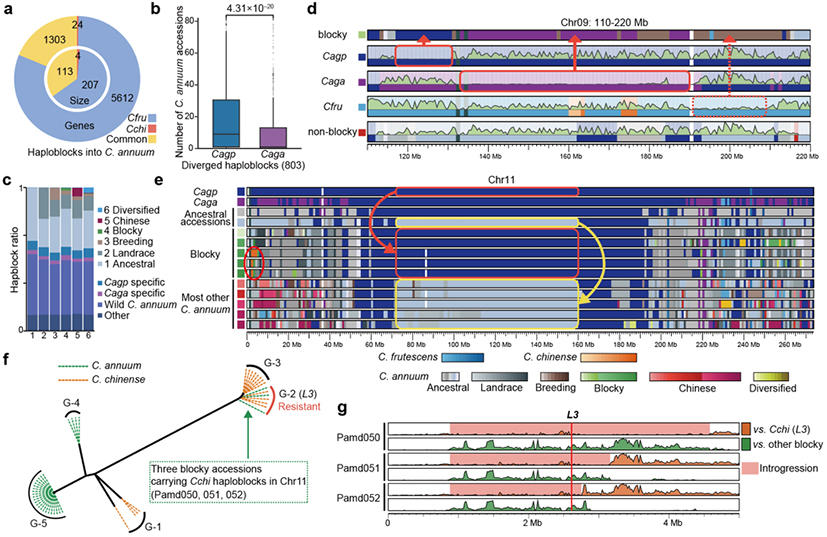
Fig 4. The pepper haploblock map and the identification of introgression events
In summary, this study, based on 11 high-quality Capsicum genomes, revealed how chromosomal rearrangements and TE amplifications contributed to the evolution of 3D genome structure and gene regulatory elements. It also highlighted the critical roles of structural variations and frequent introgressions in shaping pepper genetics and traits. These findings provide valuable insights for pepper germplasm innovation and molecular breeding strategies.
The study was mainly conducted by Dr. Zhang Kang, Dr. Yu Hailong, and PhD candidate Zhang Lingkui. Dr. Cheng Feng and Dr. Wang Lihao served as co-corresponding authors. This research marks another significant achievement of these two teams, following their work on the Capsicum variome (Molecular Plant, 2022) and the completed C. annuum genome (Plant Communications, 2024). The study was supported by the State Key Laboratory of Vegetable Biobreeding, the National Natural Science Foundation of China, the National Key Research and Development Program of China, the Innovation Program of the Chinese Academy of Agricultural Sciences, and the Central Public-interest Scientific Institution Basal Research Fund of the Chinese Academy of Agricultural Sciences.
Original article link: https://www.nature.com/articles/s41477-025-01905-1